Project description
Our best estimates show there are over 7 billion people on the planet and 300 billion stars in the Milky Way galaxy. By comparison, the adult human body contains 37 trillion cells. To determine the function and relationship among these cells is a monumental undertaking. Many areas of human health would be impacted if we better understand cellular activity. A problem with this much data is a great match for the Kaggle community.
Just as the Human Genome Project mapped the entirety of human DNA, the Human BioMolecular Atlas Program (HuBMAP) is a major endeavor. Sponsored by the National Institutes of Health (NIH), HuBMAP is working to catalyze the development of a framework for mapping the human body at a level of glomeruli functional tissue units for the first time in history. Hoping to become one of the world’s largest collaborative biological projects, HuBMAP aims to be an open map of the human body at the cellular level.
This competition, “Hacking the Kidney,” starts by mapping the human kidney at single cell resolution.
Your challenge is to detect functional tissue units (FTUs) across different tissue preparation pipelines. An FTU is defined as a “three-dimensional block of cells centered around a capillary, such that each cell in this block is within diffusion distance from any other cell in the same block” (de Bono, 2013). The goal of this competition is the implementation of a successful and robust glomeruli FTU detector.
You will also have the opportunity to present your findings to a panel of judges for additional consideration. Successful submissions will construct the tools, resources, and cell atlases needed to determine how the relationships between cells can affect the health of an individual.
Advancements in HuBMAP will accelerate the world’s understanding of the relationships between cell and tissue organization and function and human health. These datasets and insights can be used by researchers in cell and tissue anatomy, pharmaceutical companies to develop therapies, or even parents to show their children the magnitude of the human body.
Exploratory analysis
Data structure
The dataset contains information about 15 histological kidney samples. For each sample, the following features are provided.
- A tiff image of the histological sample with a resolution of 20/40k pixels in width or height
- Tabular information about the resolution of the image and the laterality of the source kidney, and also the dornor’s information: race, etnicity, sex, age, weight, height and bmi.
- The regions of the image corresponding to two kinds of tissues: Medula and cortex
- The regions of the image corresponding to the glomerula. This information is the feature to predict.
Images
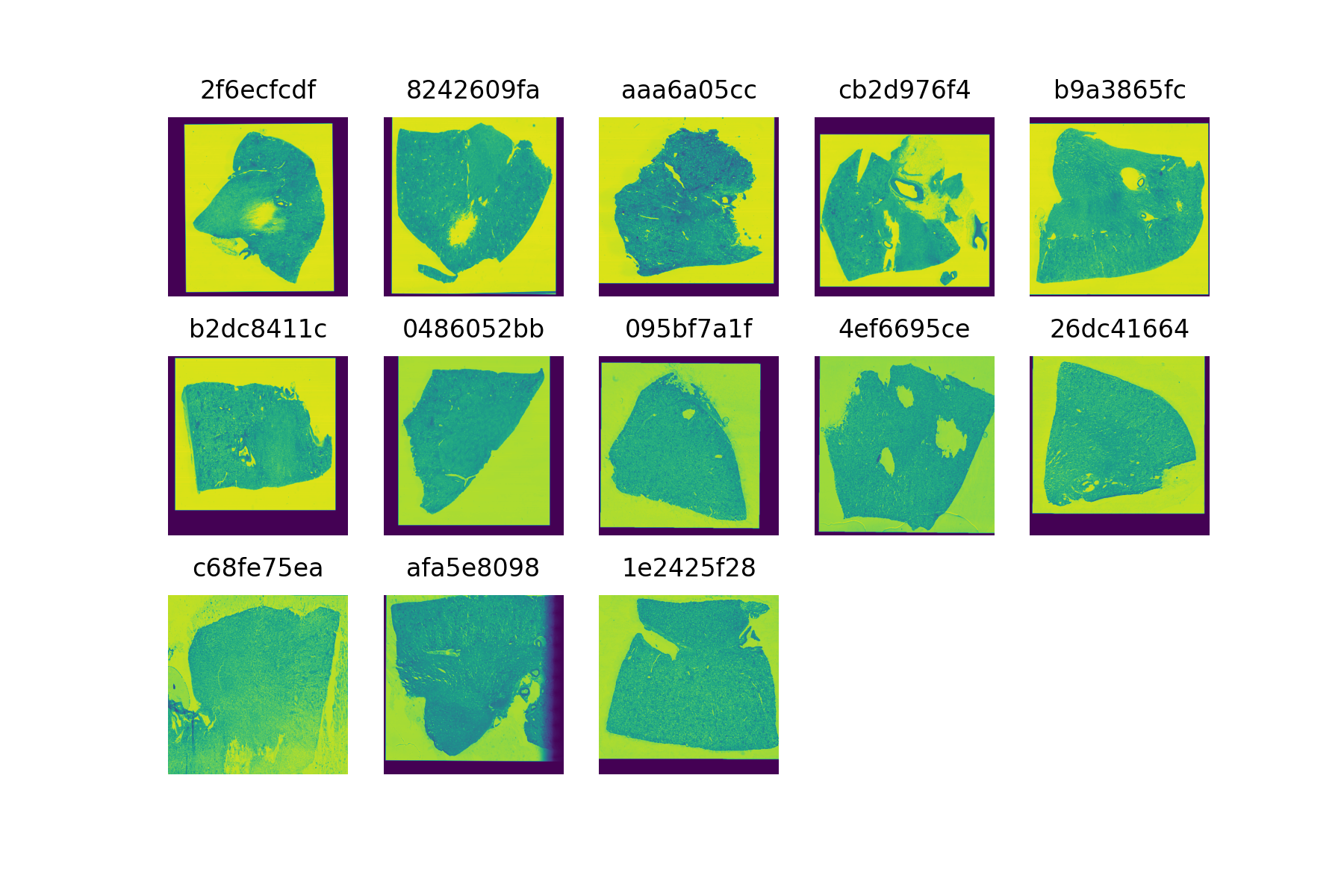
Regarding the images, several details must be noted. First two of the images are deffective and cannot be loaded (e79de561c and 54f2eec69). Therefore, the number of available samples is not 15 but 13. On the other hand, 4 images only have one channel whilst the other 9 have 3. To be able to include all the samples in the analysis, images with 3 channels will be reduced to 1. An alternative could be ignoring the 4 samples with only one channel and compare the performance of the two models. The following image shows the thumbnails and sample ids of the available samples.
The range image sizes is large, as the larger image is several times bigger than the samler one. However, any image is too large to be processed at once so this does not give us any information to rescale images. Each image will be processed as a colection of smaller images and their size will be determined by the size of the glomeruli. The following image depicts the sizes of the different images with the larger dimension in the y axis and the smaller in the x axis.

Cortex and Glomeruli
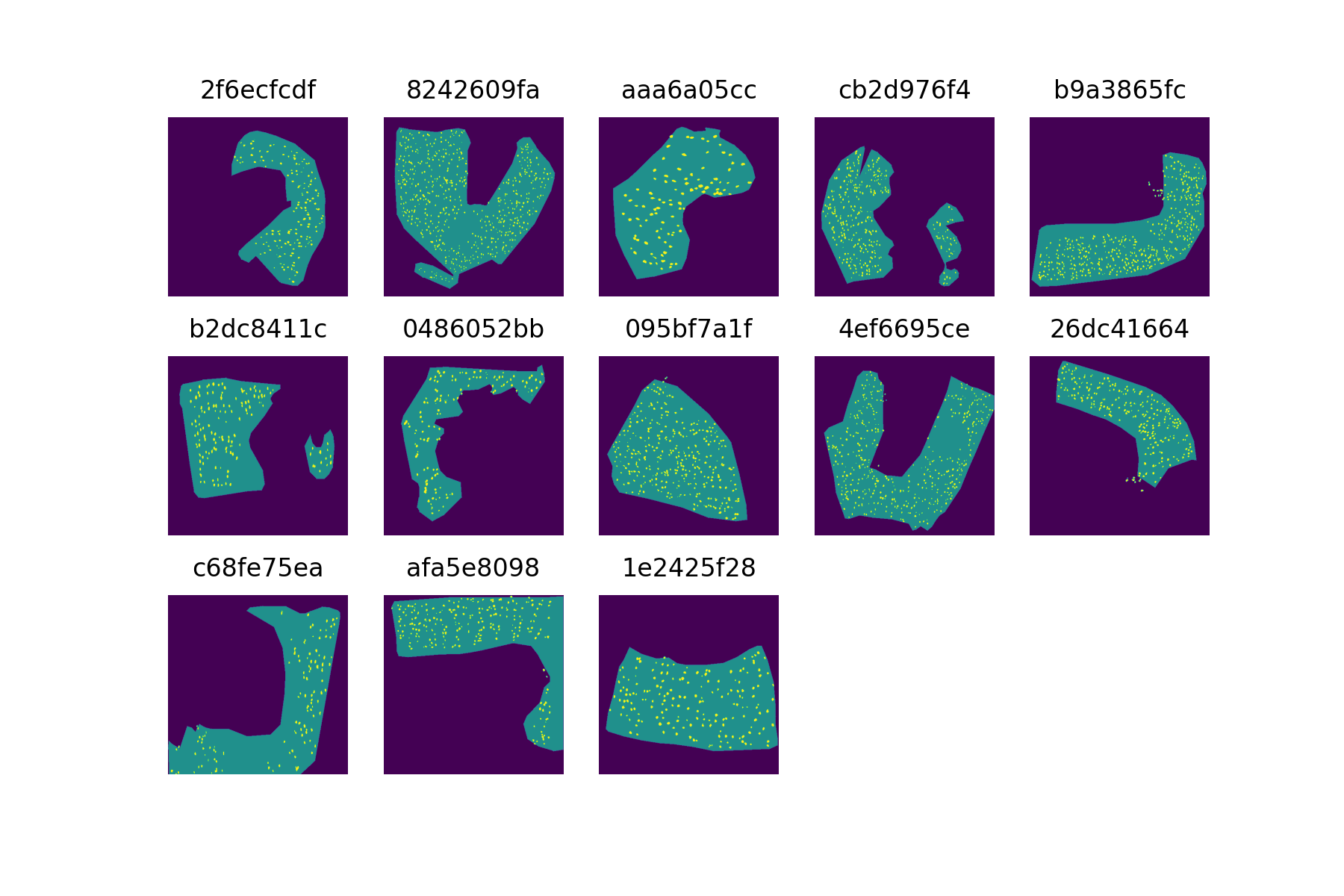
For each image we have information about three regions: the medula, the cortex and the glomerula. The medula and the cortex are the two types of tissues that can be found in the sample. The cortex on the other side, are small groups of cells scattered almost exclusively on the cortex. Because of this, the small images that we are going to use to train our model will be taken only from cortex. The next image shows the poligon marking the cortex of a sample and also the glomerula. As can be seen all glomerula fall inside the cortex of the sample.

In the next figure, we can appreciate that the same pattern repeats in the rest of the samples. In some isolated cases, glomerula can be found in the medula but it is only a small proportion of the cases.
Patient data
In this project, we need to identify the location of the glomerula by generating a mask that overlaps with them. This makes impossible the analysis of the patient data with respect to the variable of interest as we would do with a regular regression or classification. Despite of this, we can add this data to a model and evaluate if the accuracy is improved by that.
Approach
Data Preparation
As stated before, from each sample image several smaller images where generated. The image size was fixed to 2000 squared pixels and the center was randomly picked from the cortex region. The image to predict consisted in the glomerula mask corresponding to the same window region than the sample sub-image. Finally the image/glom mask pairs for each sample where saved in tfrecord. In this way, it is easier to choose which samples will form the test set and which not. Also this will avoid that imges from the same sample will end up in training and testing sets.
Model
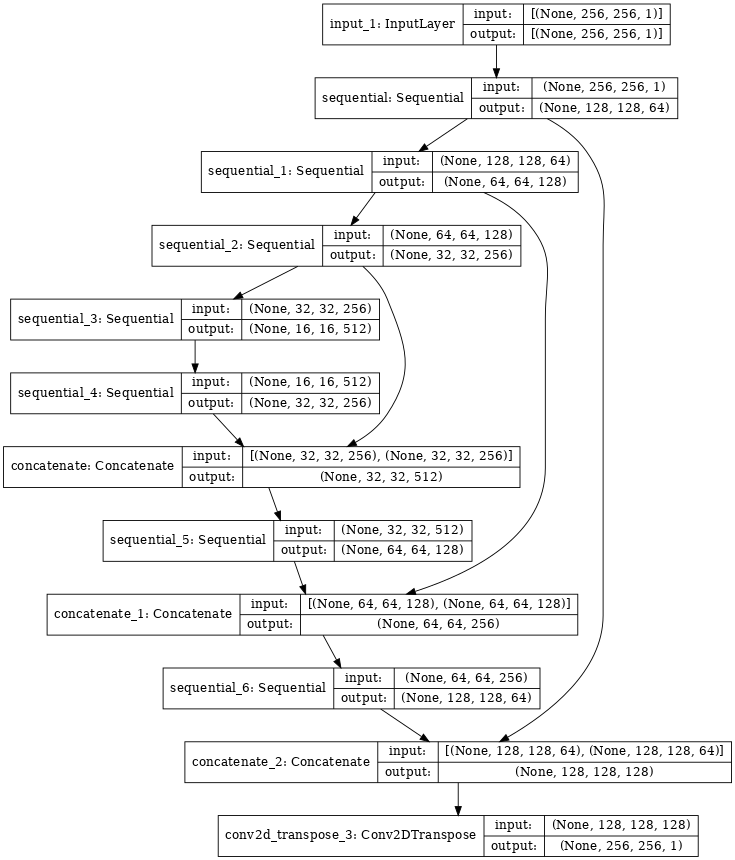
The problem is approached like an image segmentation problem. The model choosen to do this is a modified U-net . This is an Artificial neural network which input is a sample sub-image and the desired output is the corresponding region of the glomerula mask. The size of the layers reach a minimum in the center of the network, which allow to encode the original image in a summarized way. Also, non consecutive layers are connected between them.
Loss function
The used loss function is the same used for evaluation in the hubmap competition, the reversed dice coefficient. The dice coefficient is 1 when the predicted image and the target are identical and 0 when not a positive pixel of the target is predicted.
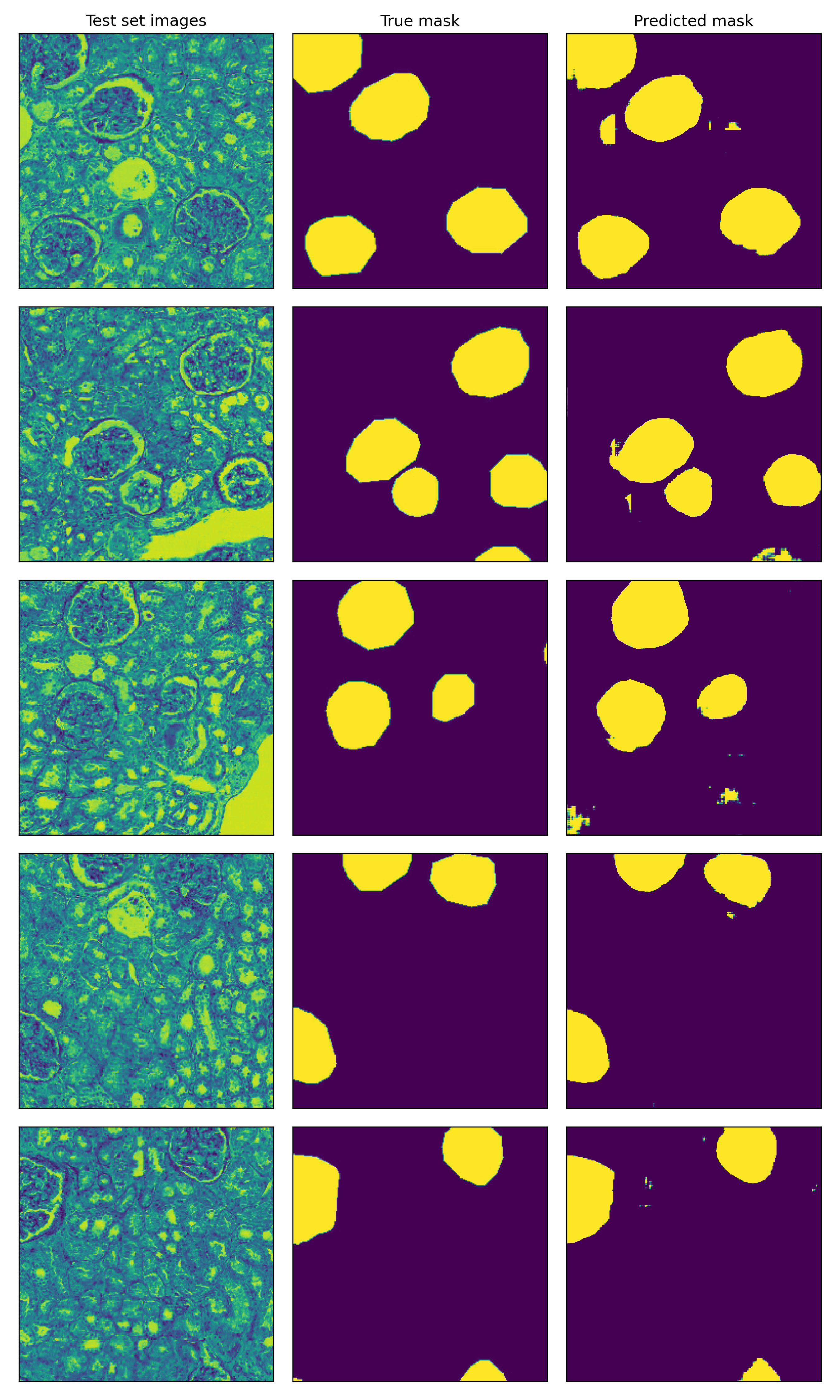
Results
The resulting model was able to achieve decent performance with a dice score around 0.4 in the testing set. Here, I show some images of the testing set, along with the true mask and the predicted mask from left to right. Although in some cases there are some false positives and background noise, the model is able to pick up almost all the glomerula in the sample of images. To pick these images, I have filtered those with at least 10% of positve pixels in the true mask.